By David M. Brady, ND, DC, CCN, DACBN, IFMCP, FACN1 and Cass Nelson-Dooley, MS2

Molecular Methods Revolutionize Diagnostic Testing
In 1885, the first microbe was cultured. In 1969, breakthroughs in anaerobic microbial culture helped further define the GI ecosystem. But in 2000, molecular techniques, or DNA analysis, sparked what was initially described as a renaissance in the laboratory and later called, the “molecular revolution.”1 These molecular methods, also used in the landmark Human Microbiome Project, made it possible to see at least 50 percent more microbes than had ever been seen before.2-4
Methods of Stool Testing Defined
Polymerase chain reaction (PCR) methodologies are DNA replication methods that make numerous copies of a target sequence of DNA in the presence of primers (short, single-stranded sequences of nucleic acids) and DNA polymerase (the DNA-replicating enzyme). PCR methods are targeted approaches for rapidly detecting, identifying, differentiating, and quantitating specific microbes and genes of clinical relevance.
- Quantitative PCR (qPCR) or real-time polymerase chain reaction (RT-PCR) is widely used in biomedical research as well as in clinical diagnostics to accurately identify and quantitate specific organisms or genes present in a sample. In qPCR, segments of DNA that are highly specific for selected targets are amplified (replicated many times). Colorimetrically labeled DNA probes (single-strand nucleic acid sequences designed to bind the amplified target gene) make it possible to quantitate the amplification process as it occurs, in real time (hence the name), yielding a quantitative DNA result.
- Standard PCR is similar to qPCR, except that it is not regarded as truly quantitative because the amplified DNA is quantified only at the final stage of the PCR process, making it impossible to determine the original true quantity of target DNA. It may instead be estimated by comparing it to a standard curve or by analyzing the quality and yield of PCR-products with gel electrophoresis.5
DNA or RNA sequencing methodologies determine the order of nucleic acids (adenine, cytosine, guanin, thymine) in a DNA molecule and count them.6 They are untargeted approaches for obtaining a general, high-level profile of the microbes or genes present in the microbiome.
Metagenomic and metatranscriptomic sequencing-based approaches provide a general profile of the microbiome. Both approaches involve high-throughput methods for sequencing nucleic acids, followed by advanced computational analyses on the resulting data to identify microbes, and either individual genes (DNA/metagenomic sequencing) or transcripts (RNA/metatranscriptomic sequencing). Metagenomic approaches can be used to identify potential microbial functions, whereas metatranscriptomic approaches can be used to analyze gene expression patterns. These approaches can provide approximate, relative levels of organisms, genes, or transcripts but are not considered sufficiently accurate for true quantitation. The accuracy of any given sequencing technology may also depend on the numbers of reads used. Read length is the number of base pairs sequenced from a DNA fragment. Sequencing depth refers to the number of times a given sequence has been read. Higher numbers of reads, often found in research settings, produce more accurate results. Lower numbers of reads, which helps minimize costs in commercial settings, produce less accurate results. This is a very important factor, since sequencing methods with inadequate sequencing depth are unlikely to detect clinically relevant organisms present at low levels. Quantitative PCR, on the other hand, excels at accurate detection of low-abundance organisms (i.e., the important “needles in the haystack”).
16S Sequencing is similar to other sequencing methods, but only a single gene, encoding 16S ribosomal RNA (16S rRNA), is sequenced. The 16S rRNA gene is common to almost all bacteria and archaea, which are bacteria-like organisms. Determining an organism’s abundance when using the 16S gene is not as accurate as other sequencing methods because the 16S gene varies widely in copy numbers per genome.
Culture and Microscopic Methodologies
- Culture + MALDI-TOF (Matrix Assisted Laser Desorption/Ionization Time-of-Flight) Mass Spectrometry relies on bacterial culture of the fecal specimen. The fecal specimen is plated with at least four growth medias under specific growth conditions to optimize microbial growth. Isolated microbial colonies are recovered and may be examined for phenotypic properties or screened with biochemical tests for specific identification. Isolated organisms are then identified using the MALDI-TOF MS, which is a proteomic method that measures ribosomal protein fingerprints of microorganisms, and they are then compared to a reference database.
- Ova and Parasite Examination (O&P, Microscopy). A routine O&P detects parasites and ova in fecal specimens using macroscopic and microscopic characteristics. Microscopic evaluation consists of a direct wet mount, concentration, and permanent-stain smear.

Fecal specimens are analyzed by a lab technician using a bright-field microscope. Accuracy is highly dependent on the expertise of the technician. Concentration methods, which are intended to increase the likelihood of finding ova, cysts, and larvae, can inadvertently reduce the numbers of cysts and ova in the sample. This may lead to underestimation of parasites in a stool specimen.7,8
Advantages of Quantitative Molecular (qPCR) Technology in Clinical Settings
qPCR is quickly becoming the standard for diagnostics due to the increased specificity, sensitivity, and reproducibility of PCR techniques. qPCR panels are able to rapidly detect and quantitate viruses, parasites, and anaerobic bacteria, which can be missed by traditional methods.9,10 Government and private institutions around the world are incorporating it as “standard of care” in the testing of the GI microbiota. The FDA has cleared multiple molecular-based gastroenteritis testing panels, often used in hospital labs and focused only on a short list of pathogenic organisms associated with acute diarrhea and GI distress.10 So, while there are many limited organism PCR diagnostic tests for gastroenteritis used in conventional medical settings for screening for common overt pathogenic organisms, an expanded panel that quantifies each pathogen and non-pathogen (including the clinically relevant commensal and opportunistic organisms) is a required tool to assess both the acute and chronic GI complaints that are commonly seen in ambulatory integrative and functional medicine clinics.
qPCR technology provides the ordering clinician with true quantitative values, versus simply a positive/negative result. It helps differentiate trace levels of an organism from frank elevations indicative of active infection. The qPCR method provides absolute values — not relative levels — of each microbe. This information gives the clinician important clinical insight that allows them to create personalized treatment plans for even the toughest cases.
Each analyte on a quantitative (qPCR) stool analysis should be individually validated and should meet or exceed federal Laboratory Developed Test (LDT) and CLIA requirements. Before adding any organism to a report, the following analyses should be completed successfully:
- Assay specificity – the assay detects the intended organism and nothing else
- Assay sensitivity – the assay can measure accurately within a certain range of detection (e.g. how low and how high the organism can be quantified)
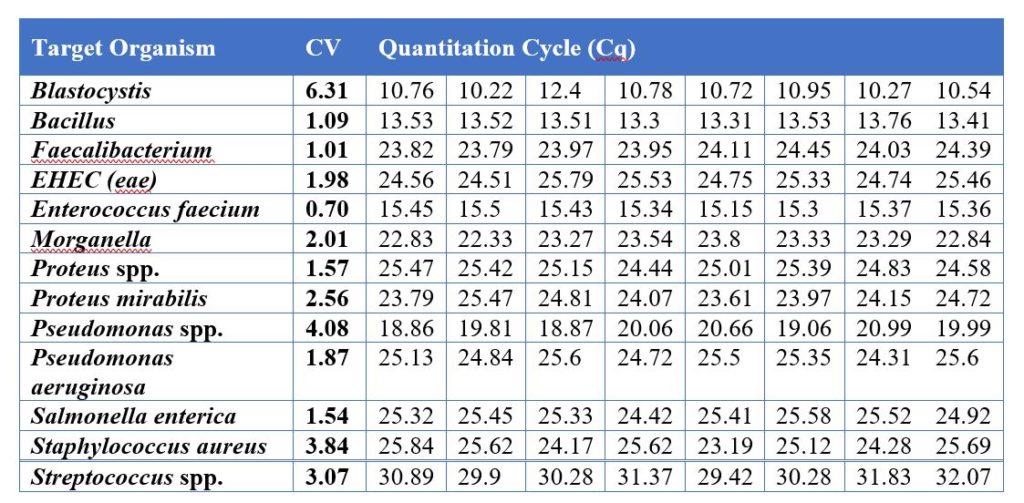
- Assay variation – if the same sample is tested multiple times, in different batches, on different days, the variation (coefficient of variation, CV) must be below 10%
- Reference range development
- Cross-assay comparison, when available.
According to CLIA standards, all organisms quantified by qPCR should have less than 15% CV or variation. This means that two identical samples, tested on different days, can only vary 15% from each other. Validation reports should be published internally, approved by laboratory leadership before adding any organism to a test panel, and reviewed during CLIA inspections. Generally, validated DNA positive controls from vendors such as ATCC are used to test molecular targets during assay validation. All lab assays and laboratory personnel should undergo proficiency testing, as required by CLIA. Proficiency testing is the analysis of unknown samples submitted by an authorized provider as a measure of external quality control.
All patient samples should be tested alongside control samples, standard samples, and endogenous controls to meet quality control requirements.
- Negative controls contain no target DNA.
- Positive controls contain a known amount of target DNA.
- Standard samples contain known concentrations of each target organism at serial dilutions. They should be run on a routine basis and used to establish a calibration curve with a coefficient of determination (R2) > 0.95.
- Endogenous controls are target organisms detected in most clinical samples.
These quality control measures allow multiple checkpoints in the assay to verify adequate nucleic acid extraction and proper amplification. If results for positive or negative controls, standard samples, or endogenous controls are abnormal, or if results are questionable for any other reason, DNA should be re-extracted, and the assay should be repeated. All patient results should be reviewed at multiple levels of laboratory management.
Discussion
The molecular revolution in stool testing is here, and it is here to stay. Culture methods have severe limitations, including within the laboratory and in sample transit. Culture methods are simply unable to truly quantitate microbes or analyze many anaerobic GI microbes at all. Within the molecular realm there are various options. DNA and RNA sequencing methods, including the most common 16S approach, offer the ability to test for a wide array of organisms at higher taxonomic level in a cost-effective manner, but lack the ability to quantitate and report to the clinician just how much of anything is actually present in a specific patient. These sequencing techniques are most appropriately used in research settings where the goal is the look across a wide array of organisms and to determine their relative populations to one another within a subject’s sample. This is why test results for 16S sequencing and metagenomic and metatranscriptomic sequencing-based methods are reported as a percentage of the total DNA found, versus an absolute quantitative amount of organismal DNA found. When the goal is to test a large number of samples within a cohort of subjects and typify the microbial composition of their guts, regardless of if the organisms found may be considered clinically significant or not, then sequencing is the logical choice. The usual intention is to then apply big-data number crunching to the resulting data-set in an attempt to determine what is typically seen in a specific population of subjects, such as those with rheumatoid arthritis, diabetes, obesity, or a host of other diseases, versus controls. Both 16S and metagenomics/metatranscriptomic sequencing methods are beneficial in a research setting when exploring the breadth of the microbiome and uncovering new species, without quantifying the exact amount present.
However, if the goal is to use the test diagnostically on an individual subject and then apply that information to make clinical decisions, then the most appropriate methodology is PCR, and a quantitative method such as qPCR is ideal and preferred. This is generally performed on a curated target list of clinically relevant organisms where the clinician is provided information not only of what is present, but how much of each organism. A laboratory’s selection of clinically relevant targets (organisms) is of utmost importance to maximize the utility of the panel in clinical practice. Further, the rapid turn-around of PCR methods is conducive to a clinical setting, whereas culture-based methods or sequencing methods can take weeks or months, respectively. A well-designed panel of qPCR targets helps to determine if an infection is present, including more acute diarrhea-producing pathogenic infections. It can also detect more subtle findings of dysbiosis of commensal and/or opportunistic organisms, which often produce chronic GI complaints of gas, bloating, distension, abdominal cramping, occasional diarrhea, and other symptoms that often lead to a diagnosis of irritable bowel syndrome (IBS) or functional bowel disorder (FBD). While all microbiological methods have merit and can be used to build on our understanding of the human microbiome, molecular methods—and especially qPCR methods—offer the practitioner the most sensitive, specific, clinically relevant, and rapid results when addressing acute and chronic gastrointestinal illness in a clinical setting.
Acknowledgements: The authors would like to thank Tom Fabian, PhD, and Heather Vatter, PhD, for their technical contributions to this article.
Relevant Author Disclosures: Dr. David M. Brady is the chief medical officer for Diagnostic Solutions Labs, LLC. Cass Nelson-Dooley, MS, is a consultant to Diagnostic Solutions Labs, LLC.
References
- Rajilic-Stojanovic M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014;38(5):996-1047.
- Wade W. Unculturable bacteria–the uncharacterized organisms that cause oral infections. Journal of the Royal Society of Medicine. 2002;95(2):81-83.
- Rappe MS, Giovannoni SJ. The uncultured microbial majority. Annual review of microbiology. 2003;57:369-394.
- Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002-5017.
- qPCR vs. Digital PCR vs. Traditional PCR. http://www.thermofisher.com/us/en/home/life-science/pcr/real-time-pcr/qpcr-education/qpcr-vs-digital-pcr-vs-traditional-pcr.html.
- Bumgarner R. Overview of DNA microarrays: types, applications, and their future. Curr Protoc Mol Biol. 2013;Chapter 22:Unit 22 21.
- Manser MM, Saez AC, Chiodini PL. Faecal Parasitology: Concentration Methodology Needs to be Better Standardised. PLoS Negl Trop Dis. 2016;10(4):e0004579.
- Garcia LS, Arrowood M, Kokoskin E, et al. Laboratory Diagnosis of Parasites from the Gastrointestinal Tract. Clinical Microbiology Reviews. 2018;31(1).
- Piralla A, Lunghi G, Ardissino G, et al. FilmArray GI panel performance for the diagnosis of acute gastroenteritis or hemorragic diarrhea. BMC Microbiol. 2017;17(1):111.
- Zhang H, Morrison S, Tang YW. Multiplex polymerase chain reaction tests for detection of pathogens associated with gastroenteritis. Clin Lab Med. 2015;35(2):461-486.

Cass Nelson-Dooley, MS, studied medicinal plants in the rainforests of Panama in 2003 as a Fulbright scholar, and then launched a career in science and natural medicine. She researched the pharmacology of medicinal plants at the University of Georgia and AptoTec, Inc, and then joined the innovators at Metametrix Clinical Laboratory. She enjoys teaching, presenting, writing, and researching how to address the underlying causes of disease, not just the symptoms. She has over a decade of experience teaching doctors about integrative and functional laboratory results. In 2013, she started Health First Consulting, LLC, a medical communications company with the mission to improve human health using the written word. She created innovative videos to improve practice efficiency and motivate patients. At Diagnostic Solutions Laboratory, she analyzes GI-MAP stool tests and develops educational tools. Ms. Nelson-Dooley is the author of Heal Your Oral Microbiome and has published case studies, book chapters, and journal articles about natural medicine, nutrition, and laboratory testing.

David M. Brady, ND, DC, CCN, DACBN, IFMCP, FACN has 30 years of experience as an integrative practitioner and over 25 years in health sciences academia. He is a licensed naturopathic medical physician in Connecticut and Vermont, is board certified in functional medicine and clinical nutrition, a fellow of the American College of Nutrition, and completed his initial clinical training as a Doctor of Chiropractic. Dr. Brady has been the chief medical officer of Designs for Health, Inc. for 17 years. He is also one of the founders of Diagnostic Solutions Labs and serves as the chief medical officer for the lab. He was the long-time vice president for health sciences and director of the Human Nutrition Institute and continues to serve as an associate professor of clinical sciences, at the University of Bridgeport in Connecticut. He has appeared on the plenary speaking panel of some of the largest and most prestigious conferences in the field, including IFM, ACAM, A4M, ACN, IHS, AANP, AIHM and many more. He is in clinical practice at Whole Body Medicine in Fairfield, Connecticut, specializing in functional, nutritional, and metabolic medicine.
NOTES
1Associate professor, University of Bridgeport, College of Health Sciences, Bridgeport, CT, (USA); chief medical officer, Diagnostic Solutions Lab, LLC (DSL) and Designs for Health, Inc. (DFH); private practice, Whole Body Medicine, Fairfield, CT (USA).
2Health First Consulting, LLC, Lilburn, GA (USA); consultant at Diagnostic Solutions Lab, LLC (DSL)