Page 1, 2
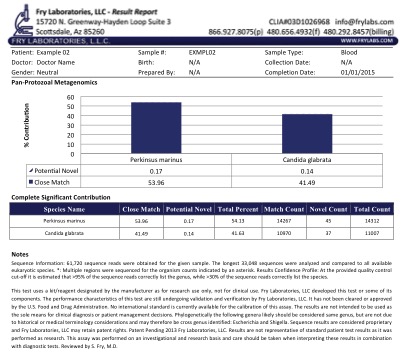
Figure 2 displays a typical result in a patient with CFS. We currently are developing a collaborative relationship with the Los Alamos National Research Laboratory investigating and documenting the presence of advanced microbes in chronic disease.
Summary
NGS systems will not replace culture systems in the near term, but it is likely that this technology will become a mainstay in the clinical microbiology arena with culture, microscopy, and serology serving as an adjunct. Eventually NGS systems will most likely replace both standard and quantitative PCR. Microscopy, serology, and emerging metabolomics will play a role in organism detection and identification; however, there is general agreement that NGS systems will dominate the infectious disease arena this decade.
Declarations
Dr. Fry owns and directs Fry Laboratories LLC, a clinical diagnostics laboratory, and is a central developer of the RIDI system.
Acknowledgements
I would like to thank Dr. Jeremy Ellis for review of this manuscript.
Notes
1. Brouqui P, Raoult D. Endocarditis due to rare and fastidious bacteria. Clin Microbiol Rev. 2001;14(1):177–207.
2. Singh S et al. Axenic culture of fastidious and intracellular bacteria. Trends Microbiol. 2012;21(2):92–99.
3. Bruneval P et al. Detection of fastidious bacteria in cardiac valves in cases of blood culture negative endocarditis. J Clin Pathol. 2001;54(3):238–240.
4. Levy PY, Fenollar F. The role of molecular diagnostics in implant-associated bone and joint infection. Clin Microbiol Infect. 2012;18(12):1168–1175.
5. Salkeld DJ et al. Disease risk & landscape attributes of tick-borne Borrelia pathogens in the San Francisco Bay Area, California. PLoS One. 2015;10(8):e0134812.
6. Biswas S, Rolain JM. Use of MALDI-TOF mass spectrometry for identification of bacteria that are difficult to culture. J Microbiol Methods. 2012;92(1):14–24.
7. Claesson MJ et al. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2012;38(22):e200.
8. Junemann, S et al. Bacterial community shift in treated periodontitis patients revealed by ion torrent 16S rRNA gene amplicon sequencing. PLoS One. 2012;7(8):e41606.
9. Pattison SH et al. Molecular detection of CF lung pathogens: Current status and future potential. J Cyst Fibros. 2012.
10. Whiteley AS et al. Microbial 16S rRNA Ion Tag and community metagenome sequencing using the Ion Torrent (PGM) Platform. J Microbiol Methods. 2012;91(1):80–88.
11. Yergeau E et al. Next-generation sequencing of microbial communities in the athabasca river and its tributaries in relation to oil sands mining activities. Appl Environ Microbiol. 2012;78(21):7626–7637.
12. Johnson G, Nolan T, Bustin SA. Real-time quantitative PCR, pathogen detection and MIQE. Methods Mol Biol. 2013;943:1–16.
13. Hedman J, Rådström P. Overcoming inhibition in real-time diagnostic PCR. Methods Mol Biol. 2013;943:17–48.
14. Wallace PS, MacKay WG. Quality in the molecular microbiology laboratory. Methods Mol Biol. 2013;943:49–79.
15. Hansen WL, Bruggeman CA, Wolffs PF. Evaluation of new preanalysis sample treatment tools and DNA isolation protocols to improve bacterial pathogen detection in whole blood. J Clin Microbiol. 2009;47(8):2629–2631.
16. Hansen WL, Bruggeman CA, Wolffs PF. Pre-analytical sample treatment and DNA extraction protocols for the detection of bacterial pathogens from whole blood. Methods Mol Biol. 2013;943:81–90.
17. Vollmer T, Kleesiek K, Dreier J. Detection of bacterial contamination in platelet concentrates using flow cytometry and real-time PCR methods. Methods Mol Biol. 2013;943:91–103.
18. Renwick L, Holmes A, Templeton K. Multiplex real-time PCR assay for the detection of meticillin-resistant Staphylococcus aureus and Panton-Valentine leukocidin from clinical samples. Methods Mol Biol. 2013;943:105–113.
19. Abdeldaim GM, Herrmann B. PCR detection of Haemophilus influenzae from respiratory specimens. Methods Mol Biol. 2013;943:115–123.
20. Abdeldaim GM et al. Detection of Haemophilus influenzae in respiratory secretions from pneumonia patients by quantitative real-time polymerase chain reaction. Diagn Microbiol Infect Dis. 2009;64(4):366–373.
21. Ling CL, McHugh TD. Rapid detection of atypical respiratory bacterial pathogens by real-time PCR. Methods Mol Biol. 2013;943:125–133.
22. Tatti, K.M. and M.L. Tondella, Utilization of multiple real-time PCR assays for the diagnosis of Bordetella spp. in clinical specimens. Methods Mol Biol. 2013;943:135–147.
23. Winchell JM, Mitchell SL. Detection of Mycoplasma pneumoniae by real-time PCR. Methods Mol Biol. 2013;943:149–158.
24. Fillaux J, Berry A. Real-time PCR assay for the diagnosis of Pneumocystis jirovecii pneumonia. Methods Mol Biol. 2013;943:159–170.
25. Yam WC, Siu KH. Rapid identification of mycobacteria and rapid detection of drug resistance in Mycobacterium tuberculosis in cultured isolates and in respiratory specimens. Methods Mol Biol. 2013;943:171–199.
26. Lavender CJ, Fyfe JA. Direct detection of Mycobacterium ulcerans in clinical specimens and environmental samples. Methods Mol Biol. 2013;943:201–216.
27. Bergmans AM, Rossen JW. Detection of Bartonella spp. DNA in clinical specimens using an internally controlled real-time PCR assay. Methods Mol Biol. 2013;943:217–228.
28. McKechnie ML, Kong F, Gilbert GL. Simultaneous direct identification of genital microorganisms in voided urine using multiplex PCR-based reverse line blot assays. Methods Mol Biol. 2013;943:229–245.
29. Van den Berg RJ, Bakker D, Kuijper EJ. Diagnosis of Clostridium difficile infection using real-time PCR. Methods Mol Biol. 2013;943:247–256.
30. Stoddard RA. Detection of pathogenic Leptospira spp. through real-time PCR (qPCR) targeting the LipL32 gene. Methods Mol Biol. 2013;943:257–266.
31. Yamazaki W. Sensitive and rapid detection of Campylobacter jejuni and Campylobacter coli using loop-mediated isothermal amplification. Methods Mol Biol. 2013;943:267–277.
32. Rimbara E, Sasatsu M, Graham DY. PCR detection of Helicobacter pylori in clinical samples. Methods Mol Biol. 2013;943:279–287.
33. Maheux AF, Bissonnette L, Bergeron MG. Rapid detection of the Escherichia coli genospecies in water by conventional and real-time PCR. Methods Mol Biol. 2013;943:289–305.
34. Barletta F, Ochoa TJ, Cleary TG. Multiplex real-time PCR (MRT-PCR) for diarrheagenic. Methods Mol Biol. 2013;943:307–314.
Page 1, 2 |
![]()
![]()
![]()





